Our Mission
The Hippo Signaling Pathway
Our lab focuses on delineating the genetic pathways that regulate organ development and homeostasis. The Hippo signaling pathway is a fundamental organ size control pathway that we first reported as an inhibitor of cardiomyocyte proliferation to limit heart size during mammalian development. As shown on the left, Hippo is an intracellular kinase cascade that signals the phosphorylation of YAP/TAZ transcriptional co-activators. YAP/TAZ are major downstream Hippo signaling effectors that partner with transcription factors, such as Tead, to regulate gene expression. Upon phosphorylation, Yap and Taz are excluded from the nucleus and rendered transcriptionally inactive. Other signals, such as mechanical signaling, regulate Yap subcellular localization. Our lab also observed that 1) inactivation of Hippo in adult cardiomyocytes unleashes productive heart regeneration and promotes reversal of heart failure and that 2) Hippo functions as a key regulator in cardiac fibroblasts and inflammatory responses. Our ongoing studies (below) are aimed at translating this work into meaningful cardiac therapy approaches.
Congenital Heart Disease
Echocardiography of control (left) and Yap mutant (right) hearts
Congenital heart disease (CHD) is the most common birth defect, arising in 10 out of every 1000 births. CHDs commonly produce pediatric heart failure (PHF), a devastating condition in which the heart has poor pumping function, leading to organ failure and death at an early age. CHD-related PHF is the leading cause of death in an infant, child, or adolescent and the associated treatment costs are exorbitant. Although current PHF treatments such as Left Ventricular Assist Device implantation and heart transplantation increase the survival rates for pediatric CHD patients, they have obvious limitations and thus, new approaches are clearly required. As such, a considerable effort has been made to develop therapies aimed at stimulating endogenous repair of cardiac muscle in PHF patients. Excitingly, these therapies, while currently unavailable and long considered to be an impossible goal, are now within reach. CHD is multigenic, and the genomic properties of PHF patients are poorly understood. Hence, our goal is to identify the biomarkers that can be used to categorize PHF patients to determine the genetic programs that can be manipulated to increase endogenous cardiomyocyte renewal and aid in repairing the heart. These studies will yield important insights of the molecular mechanisms of CHDs, will refine HLHS and CHD therapeutic targets, and have an enormous potential to innovate PHF diagnostics and treatment of PHF and other cardiac diseases.
Cardiac Regeneration and Heart Failure
Pig heart tissue after myocardial infarction: Green shows delivery of our gene therapy product to the infarct.
Heart failure due to cardiomyocyte loss and pump dysfunction after ischemic heart disease is the leading cause of death in the United States. We strive to facilitate cardiac regeneration after heart damage to improve survival rates. Limited endogenous adult cardiomyocyte regenerative potential after acute damage results from inadequate adult cardiomyocyte proliferative capacity. Strikingly, the mammalian heart endogenously replaces merely 1% of cardiomyocytes per year. In our previous work, deletion of the Hippo component Sav in mouse and pig cardiomyocytes with established ischemic cardiomyopathy and heart failure (HF) reverses HF and improves heart function due to increased nuclear YAP and activation of YAP target genes. In addition, we discovered that the dystrophin-glycoprotein complex sequesters YAP in adult cardiomyocytes. Hence, our ongoing objective is to unveil the mechanism(s) of Hippo-mediated cardiac cell regeneration. One of these mechanisms is to regulate cytoskeletal remodeling and protrusion formation. We also found that Yap makes chromatin more accessible for the expression of fetal genes, thereby promoting reprogramming of somatic cells into a primitive, fetal-like proliferative state. We work extensively with ischemia-reperfusion–induced myocardial infarction mouse and pig models and recently demonstrated that locally delivered gene therapy to inhibit Hippo in border zone cardiomyocytes is the first example of bona fide tissue renewal therapy. These studies have improved the understanding of the regulatory mechanisms that control cardiac development, disease and regeneration with the long-term goal of developing novel HF therapies.
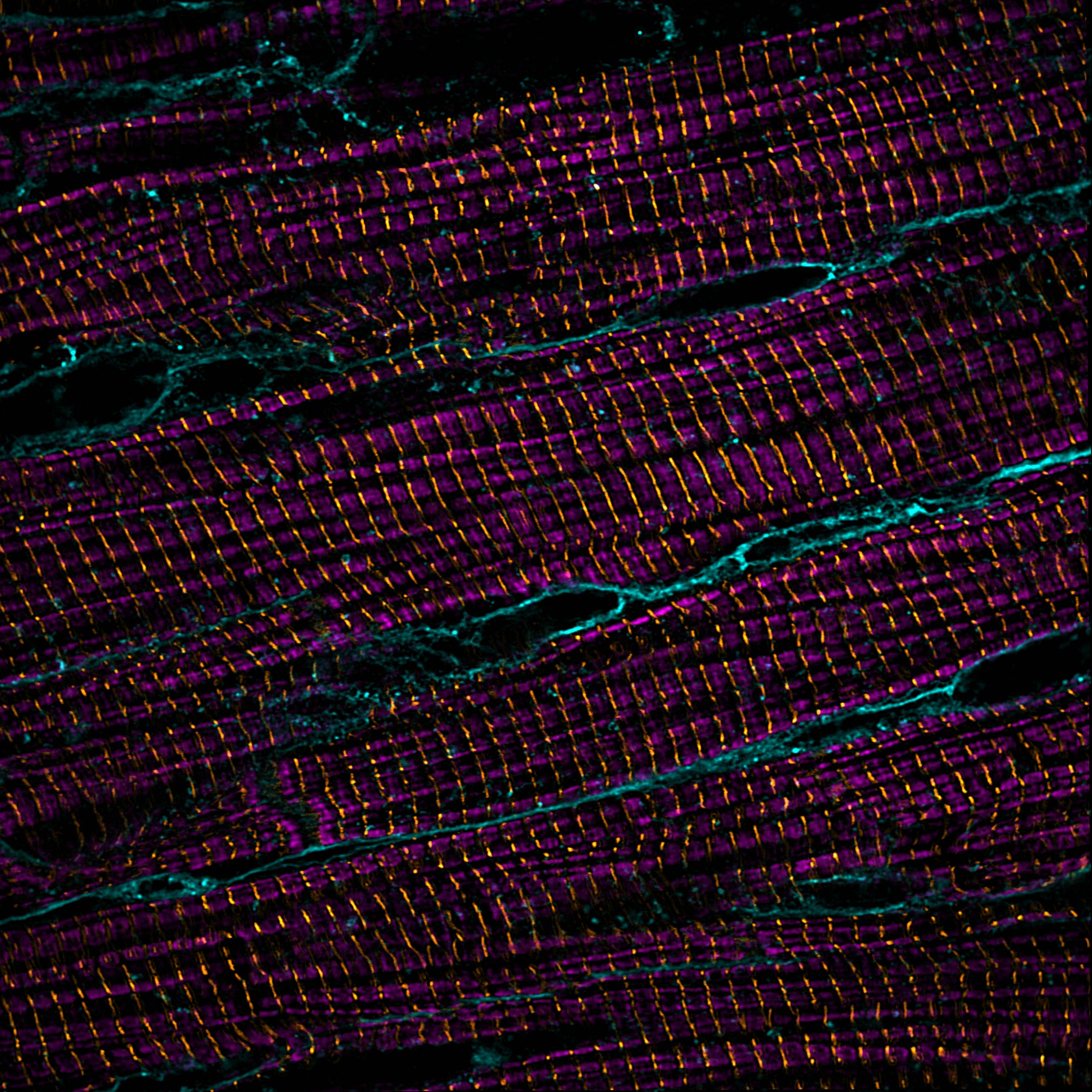
Super-resolution imaging of cardiomyoctes: sarcomere thin filaments (magenta), z-discs (yellow), cell membranes (teal)
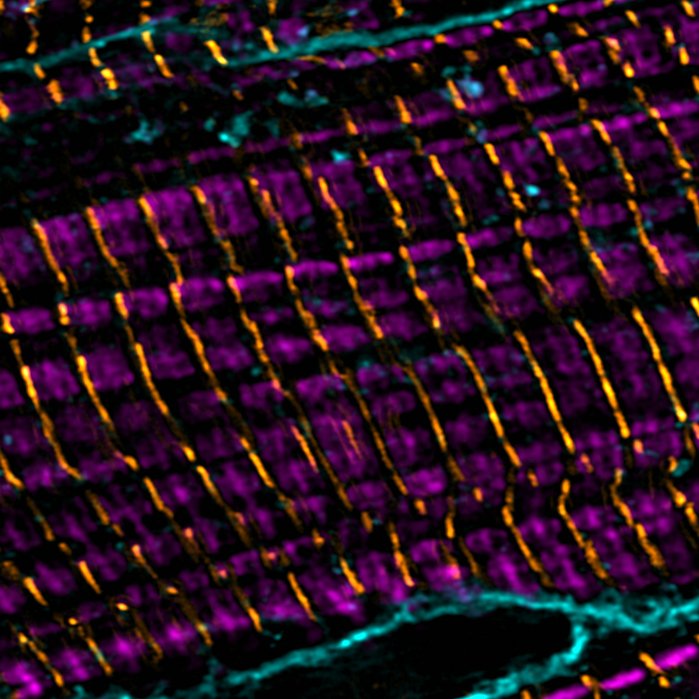
Super-resolution imaging of cardiomyoctes: sarcomere thin filaments (magenta), z-discs (yellow), cell membranes (teal)
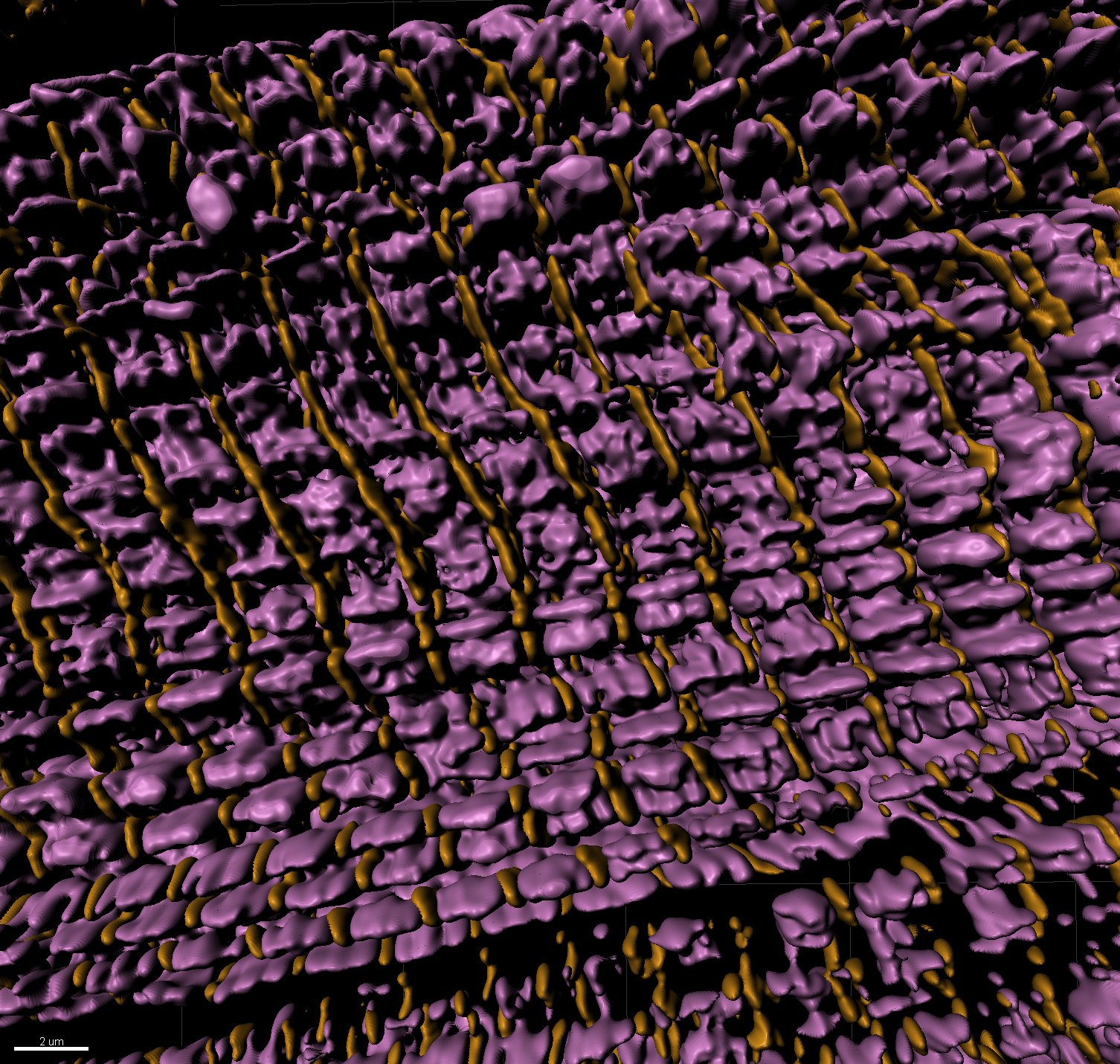
3D rendering of super resolution image: sarcomere thin filaments (magenta), z-discs (yellow)
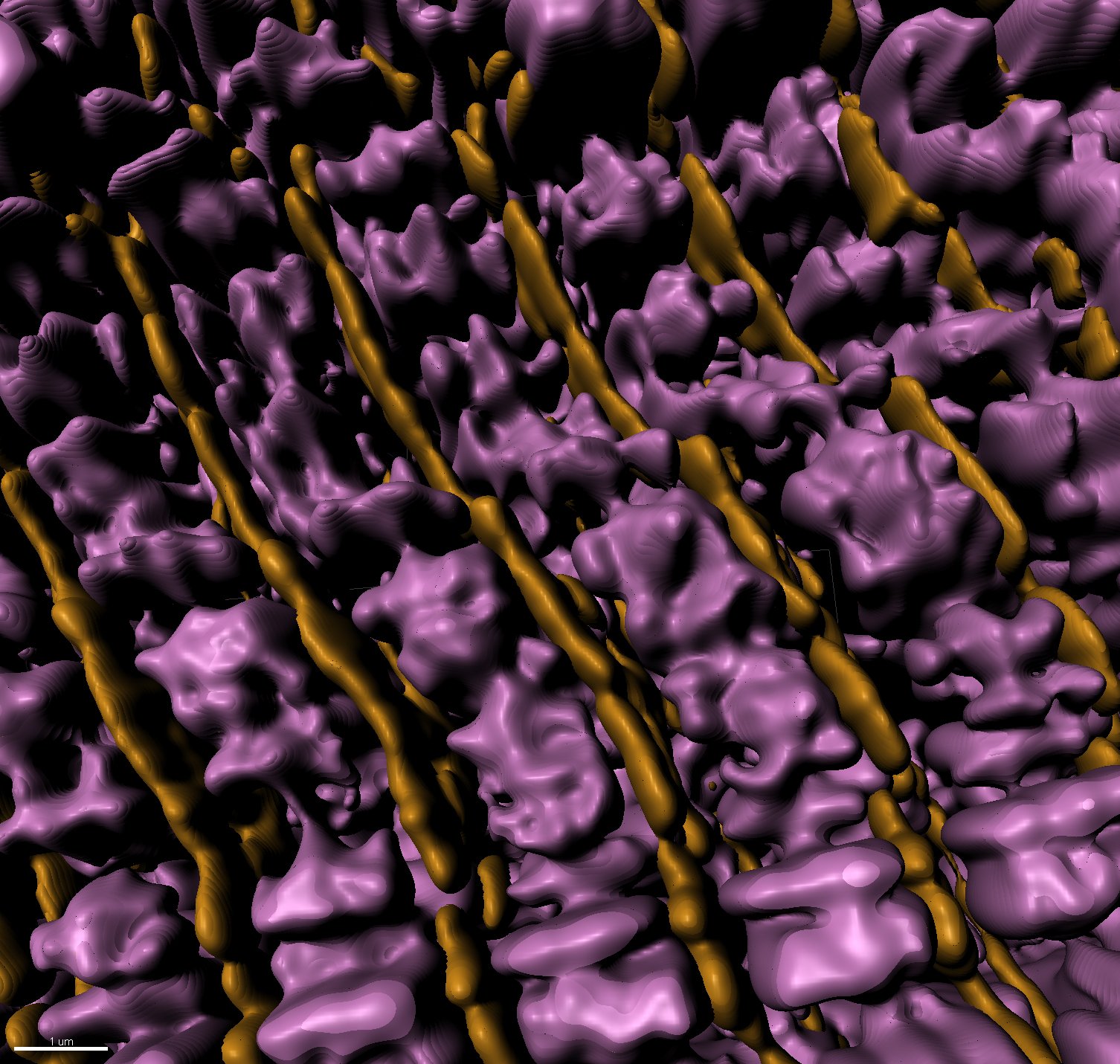
3D rendering of super resolution image: sarcomere thin filaments (magenta), z-discs (yellow)
Single Cell Genomics
Single cell mouse compendium showing diverse cell types (colored clusters) in the heart. 3D uniform manifold projection of 1 million cells and nuclei delineating 20 mouse cardiac cell types.
We have a long-term interest in studying transcription factors involved in cell fate transitions in development and regeneration. Our lab has extensively studied transcription factors, initially using mouse models and reporter alleles, which include paired-related homeobox genes. Over the past seven years, we have transitioned into single-cell biology and computational biology and have published several high impact papers that address an array of important biologic questions by using single-cell multi-omics approaches.
To this end, we are constructing a compendium containing one million cardiac single cell transcriptomes that provides a comprehensive set of snapshots of the homeostatic cellular landscape of the mouse heart and spans from embryonic stage E7.5 to 2.5 years of age. A portion of this compendium comprises cells obtained after ischemic injury, ranging from one to ninety days post-injury. Through these efforts, we have annotated over 20 cell types present in the heart and we have thus far resolved dozens of distinct cell states for each of the cell types. Altogether, our compendium will provide a powerful and highly rigorous tool for data-driven discoveries.
Differential expression of genes in the heart visualized by spatial transcriptomics